Genome sequencing is coming to your home
Our human DNA comprises of 20,000 to 25,000 genes. An organism’s DNA is made up of an order of DNA nucleotides, which are represented in the form of genetic letters. The human genome is made up of over 3 billion of these genetic letters.
![]()
Figuring out of this order of DNA nucleotides is very important because this genomic data unmasks many things such as
- Tells disease risk factors
- How you might respond to certain drugs
- The traits that you might pass down to your children
- Genome sequencing also allows the identification of signatures of host adaptation, identification and monitoring of diagnostic targets, and characterization of responses to vaccines and treatments.
Today, DNA sequencing is on a large scale. It uses high-tech machines, which read the sequence of DNA bases and translates from life’s chemical alphabet into our alphabet. But it is not that simple. The entire process takes months of time and costs about six figures to read the entire Genome. This cannot be suitable for all conditions because
- It ‘s hard to transport laboratory equipment for some rural areas
- It is impossible to maintain skilled human operators everywhere
- The time taking is too long, as some diseases will show rapid reaction on human body
- The cost is prohibitive that some sectors of people cannot bear it.
So, there is a need for miniaturization of DNA sequencers and reduction in their time and cost. If I say that, the entire genome sequencing equipment is coming to the size of a USB memory stick and costs around $1000, then what are you thinking? Incredibly small, isn’t it?
What is Portable Genome Sequencing?
The process of mapping a genome by decoding all 3 billion letters in human DNA is called Genome Sequencing. It is said to be done with the help of individual machines which scans the sequence of letters like a human eye. This seems to be not suitable for all conditions in the world. So, Oxford Nanopore has been developing a disruptive nanopore-based technology for sequencing DNA, RNA, Proteins and other long-chain molecules. It is called as MinION. It is about the size of a USB memory stick and obtains both power and computer analysis from a standard laptop computer. It is capable of sequencing 100 million base pairs within its six-hour working life. The device accepts samples of whole blood, plasma, and serum for quick analysis and the reagents required for the analytic process come ready to use with each MinION.
“At first, the idea of using a nanopore to measure DNA was a bit like Jules Verne writing about flying a rocket to the Moon. Fine, but how?” Clive Brown
| Recommended for you | |
| Oxford Nanopore’s MinION: A portable genome sequencing device | |
| Preventive medicine in the omics era | |
| Optogenetics cure blindness and parkinsons’ disease |
How does it work?
A nanopore is an organic molecule penetrated by a very small (i.e., 1-2 nanometre) hole. These nanopores are enclosed in a synthetic polymer membrane which is only two molecular layers in thickness. Initially, bilayer lipid membranes, similar to those resembling throughout the body, were employed but proved far too fragile to handle in a production sequence. Development of such a thin layer that has considerable electrical resistance and is sufficiently rugged for use in the field was a significant ON project.
A nanopore is positioned upon such a membrane so that the hole in the nanopore traces a path from one plane of the membrane to the other. Later, the layer is then positioned so that it separates a volume of an electrophysiological fluid in half.
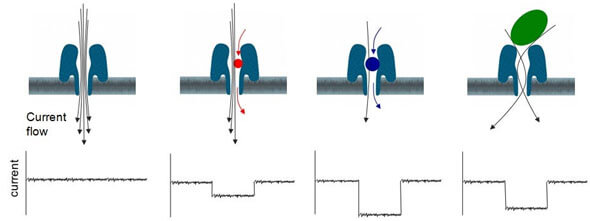
The image above shows a nanopore enclosed in a layer. A voltage difference is placed between the two halves of the liquid. The liquid contains free ions, so an ion current then moves through the centre of the nanopore. This ionic current acts to clear nearby molecules, proteins, and even RNA or DNA into the mouth of the nanopore.
If a target molecule is brought into or upon the nanopore, it disrupts the flow of ions in a particular manner which can be detected and interpreted.
A wide range of molecules can be distinguished and identified using the nanopore ionic current measurements, varying from small physiological or pharmaceutical molecules to proteins. Still, the MinION is directed toward DNA sequencing, so the nanopore and enzymes in the electrophysiological fluid are optimized to distinguish the four amino acids (thymine, guanine, cytosine, and adenine) which encode genes in DNA. In the final event, three amino acids are read together by the nanopore, which possesses the delicacy to discriminate between the 64 possible combinations.
The DNA strand is attracted to an enzyme that is part of the electrophysiological fluid. In turn, the enzyme is drawn to a nanopore, in which it resides. The protein is engineered to unzip the double-helix structure of the DNA strand so that one strand is addressed back into the fluid, while the other starts the nanopore for sequencing. More unusual is that the enzyme incorporates a ratcheting structure, so the single DNA strand does not move quickly through the nanopore, but rather elevates one base pair at a time. The rate at which the DNA strand passes through the nanohole is regulated by the magnitude of the unblocked ionic current and can be as high as 15 base pairs per second. Better efficiency is associated with slower rates.
Rapid development
The need for the miniaturization had made this technology to grow faster. The nanopore, which is related to the extremely small, i.e., nanotechnology, is available due to the ongoing researches in the field of Nanotechnology. Another one is that the massive challenging competition in the market. Majorly, the increase in the effectiveness of diseases causing microbes day-by-day led to this rapid technological improvement.
Privacy of our information
As this is an upcoming technology, the scientists have not taken any measures regarding privacy. So, the information that we shared may not be extremely confidential. The President’s Bioethics Commission released a statement addressing the uneasiness around the topic.
The report contained recommendations urging federal and state governments to develop processes to protect the privacy of persons interested in retrieving genomic data.
But while the public records concern for privacy around the issue of genome sequencing, experts in the industry, along with Merel, don’t seem bothered. For him, genomic data will one day be considered similar to having banking information on one’s computing device.
Green doesn’t see privacy as an immediate issue either but does admit that with the rise of genetic data out there, the opportunity develops for it to land in the hands of someone who shouldn’t be looking.
What advantages does it offer?
After years of technological improvements and researcher’s hard work, genome sequencing can be done in days instead of months. The cost instead of paying an amount of six figures, pay as little as, $99 for a limited view of their data and around $9500 for their full genome. This technology offers a broad range of advantages for the customers and reduces the burden on medical operators.
- Real-time DNA/RNA sequencing
- Ultra long reads
- 10 minutes library preparation
- Scalable to portable (or) desktop
- It can be employed in many areas of research
- Pathogens/ Microbial/ Antimicrobial resistance
- Environmental Research
- Microbiome
- Cancer research
- Human Genetics
- Basic genome research
- Plant research
- Clinical research
- Transcriptome analysis
- Population Genomic
- It can deliver a range of biological analysis techniques
- Whole genome sequencing
- Targeted sequencing
- RNA sequencing
- Metagenomics
- Epigenetics
- It can diagnose infectious as well as hereditary diseases
- High fidelity
- No capital cost is required
- There is no need for skilled medical operators
- No PCR or other amplification technique is required
- Whole blood, plasma, and serum can be accepted
- It can be used to sequence animal genome also but not only the human ones
Applications
It is suitable for many applications
- Water testing
- Food safety and efficiency
- Infectious disease and microbiology
- Environmental monitoring
- Biodefense/ outbreak surveillance
- Supply chain control and authentication
- Agricultural: both plant and animal
- Forensics
- Industrial diagnostics
- Pharmaceuticals
- Oncology
- Clinical genetics
- Reproductive medicine
- Consumer genetics
- Education
Challenges
As this technology is awe-inspiring and offers a broad range of advantages and applications, there are also particular problems which are to be fulfilled by the team
- To reduce the cost for less than $1000 or even $100 for full genome sequence
- To facilitate the analysis of any living thing, by any person, in any environment
- To increase the market of genome sequencing from developed countries to very rural areas
- Acceleration of technology and better use of computer to make the entire process user-friendly
A lot of people compare the genomic testing revolution to the rise of the personal computer. There have been many ways we’ve used the personal computer, all of which had gone past anything that could have been imagined meanwhile Steve Jobs was creating the first Macintosh.
In spite of the potential miniaturization of the DNA-sequencing platforms, biological samples that comprise DNA material to be sequenced may have to undergo a special treatment. Sample-preparation activities including extraction and PCR amplification of the DNA samples to be sequenced which may require the hands-on experiences of a trained person and thus put DNA sequencers beyond the reach of ordinary consumers. For long-term storage, DNA-sequencing reagents, including enzymes and nucleotides which require refrigeration to avoid spoilage. Such factors may prevent DNA-sequencing technology from leaving the confines of air-conditioned laboratories in the hands of highly skilled individuals and may need to be addressed before DNA-sequencing biosensors can become widely used in commercial bio-sensing applications.
Companies providing these sequencers
- Ion Torrent has announced the full genome sequencing for just $1000 or less with its sequencing platform, Ion Proton sequencer. It is the world’s first desktop semiconductor-based gene sequencer. It is also the first machine that can read genome in two hours.
- 23andMe offers one of the most successful direct-to-consumer options. For $99, a consumer can order a testing kit, spit into a tube and give it back to the firm. From there, 23andMe will give the sample to a lab and get back with a genotype record. Genotypes vary from genomic sequences in that the data provides patient’s access to around 1 million genomic “markers.” A full series includes 3 billion letters, giving consumers a complete view. But 23andMe’s less expensive offering provides more than 240 personalized genetic reports on how you will react to medication, your risk for specific diseases and physical traits, Afarian says. Couples can learn the probability of traits in their offspring when both parties get tested and use the company’s Inheritance Calculator. The company also released an Android app titled 23andMe Mobile where consumers who’ve already done the DNA test can view results.
- Another one is from San Diego, California. Illumina announced Hiseq 2500, a “genome-in-a-day” sequencer. Its cost of sequencing each genome is between $3000 to $5000, depending on the number of samples that are to be sequenced. Illumina also came out with MyGenome, an iPad app that allows users to interpret their entire genome graphically.
- Also in San Diego is Portable Genomics, a first stage software company slated to issue a digital product in the coming few years that will enable consumers to analyze genomic data. The product, says founder Dr. Patrick Merel, a little diagnostic expert, will offer a graphical interface that will direct patients to nearby specialists and provide medical testing reminders. The biggest draw will be the cost: $100.
- Knome has developed exclusive technologies that automate many of the manual tasks involved in interpreting whole human genomes. For pharmaceutical, academic, and medical researchers, Knome offers:
knoSYS® Platform. It provides labs with a scalable, production-grade informatics system for the interpretation, analysis, and annotation of human next-gen sequence data (exome, genome, targeted gene).
knome DISCOVERY Research services include project-driven curation, sophisticated informatics, and in-depth analysis by Knome’s team of experienced geneticists. - Finally, here comes the Oxford Nanopore’s MinION is a genome sequencer which is portable, i.e., possibly handheld. It was in size of a USB stick and weighing about 100 grams. Some samples can be diagnosed, and the results can be obtained in less than 15 minutes. Now, more than 1,000 researchers from 63 countries are going to apply the MinION to a wide variety of problems.
Future
Up to now, the genome sequences are limited to the developed countries. But for the countries like West Africa where Ebola virus disease is epidemic and is responsible for over 28,599 cases and more than 11,299 deaths, this genome sequencing is a nightmare. It is equally as beneficial for farmers who want to sequence their crops as it is for doctors in West Africa who need to diagnose Ebola as quickly as possible. Genome sequencing in viral outbreaks is desirable to characterize the infectious agent and determine its evolutionary rate. It is also equally important for the schools and universities. So, portable genome sequencing plays a key role in this type of cases due to its unique advantages of less time, less cost and no need for a skilled operator. So, this technology would acquire a bright future in both developed and as well as developing countries and may transform DNA sequencing into an enabling technology for biosensors.
As the technology accelerates and prices come down, Dr. Robert Green, a genetics professor at Harvard Medical School, says genomic testing will soon go mass market. And from there, anything can happen.
According to Wall street journal, government and research centres are the primary buyers for these sequencing machines. There are only about 4000 labs in the world having an Illumina machine. Then, what about the remaining? They have to adopt this type of portable genome sequencing for the better performance and reliability. So, a near future is going to bring us a technology where machines can help doctors both prevent and treat illness. Are you one of those people who think that?
Image credit: www.istockphoto.com

















I’ve heard of next-generation sequencing in the clinical study of infectious diseases, but I hadn’t heard of the environmental research that is also being done. It’s fascinating that genome sequencing can tell us so much about such a variety of topics. This makes me wonder if there are different genome markers for different types of infectious diseases or if you have one marker that makes you susceptible to a lot of them.